Methods
Currently, the methods evaluated in (Hücker et al., 2021) start with micro-manipulation to separate single-cells and perform library preparation for the detection of miRNA. Unlike mRNA, miRNA have no Poly-A tails and therefore conventional library preparation methods of single-cell miRNA preparation are not applicable. There are efforts underway to optimize the library preparation methods by manipulating different technical conditions such as buffer concentrations, enzymes used for ligation and the adapters.
For now the Sandberg Protocol with CleanTag adapters appear to give the best results in terms of miRNA capture and reproducibility across different cell culture types (Hücker et al., 2021). However, we offer the option to process alternative adapter configurations as listed below.
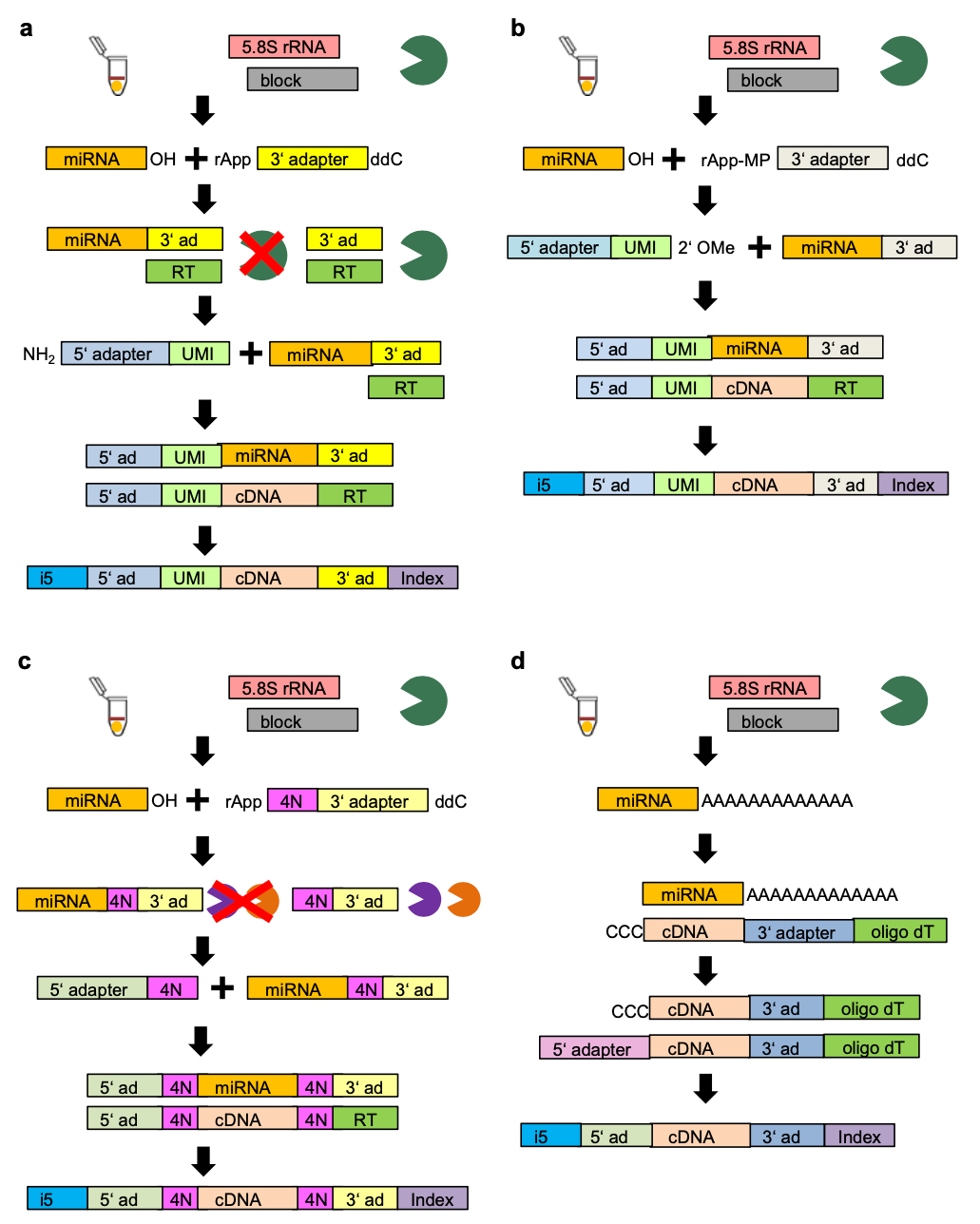
The conditions that are observed at the sequencing level, and are therefore relevant to the preprocessing of single-cell miRNA data are the UMI_length, 3’ and 5’ adapters along with the method. * Index_number is also included as a parameter, however this is the conventional Illumina TruSeq index primer and is not expected to play a major part in the performance of single cell miRNA sequencing. The protocols primarily used here (and denoted by Hücker et al., 2021) are Small-Seq protocol denoted SB and SBN (Faridani et al., 2016, Hagemann-Jensen et al., 2018), CleanTag protocol, denoted as CL (Shore et al., 2016), 4N protocol, denoted as 4N (Giraldez et al., 2018) and CATS, denoted as CATS (Turchinovich et al., 2014). A graphical representation of the structure of the reads can be found in Figure 1. Since it is possible to swap the 3’ and 5’ adapters between the different methods, we allow these methods in the corresponding columns.
Settings
Index number (Index number):
Corresponds to TruSeq adapters 1 to 12. Currently, library preparation for Illumina Sequencing as per Hücker et al., 2021 is supported. The index number has to be specified for each file in the metadata sheet as an extra column which has to be provided by the user.
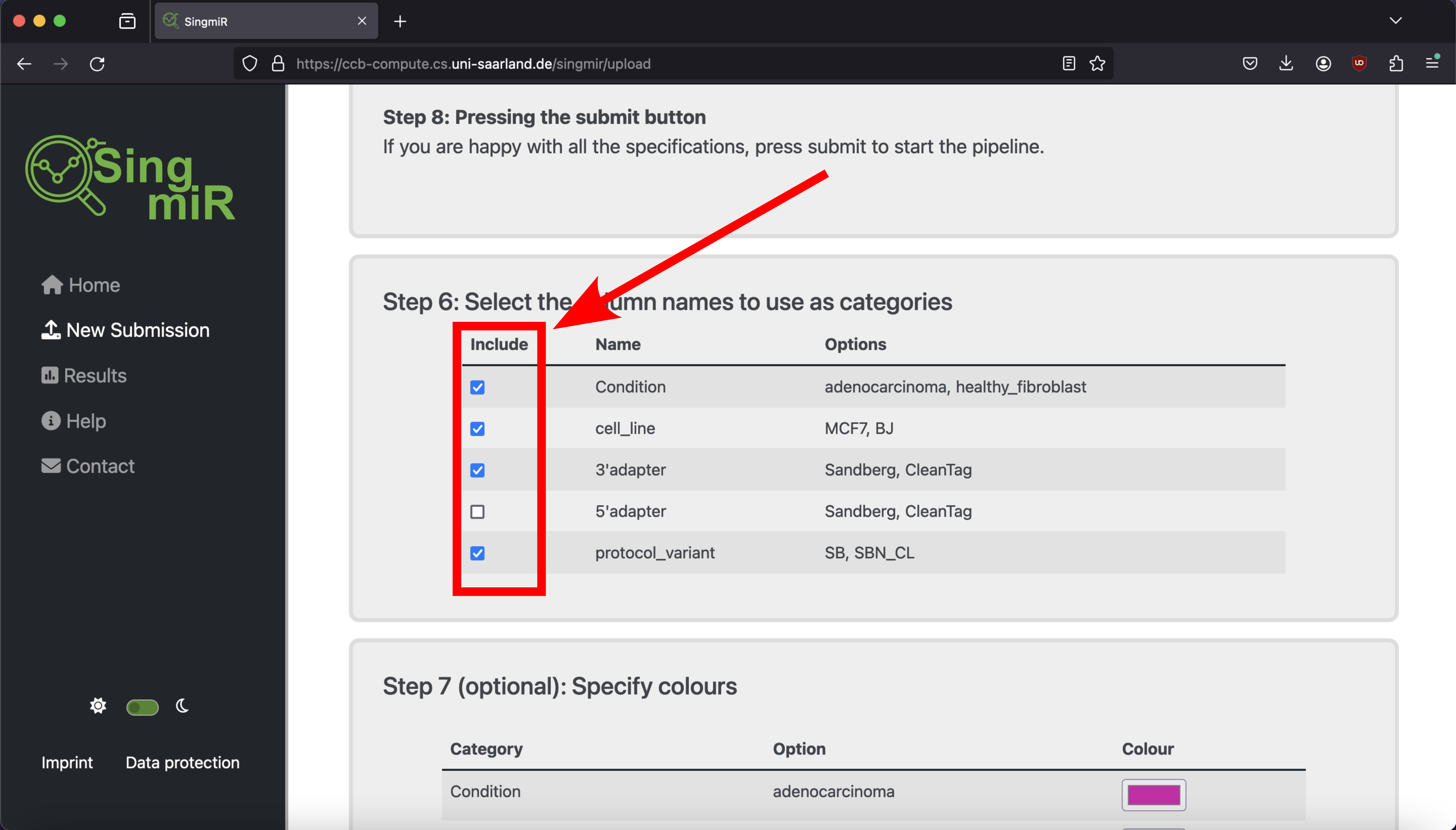
Protocol variant (protocol_variant):
For each sample a protocol variant is needed. The protocol variants can be the same for all files, therefore, we included the global parameter option. The user can also specify individual protocol variants for each file in the metadata sheet as an extra column. Parameters are separately explained under the table.
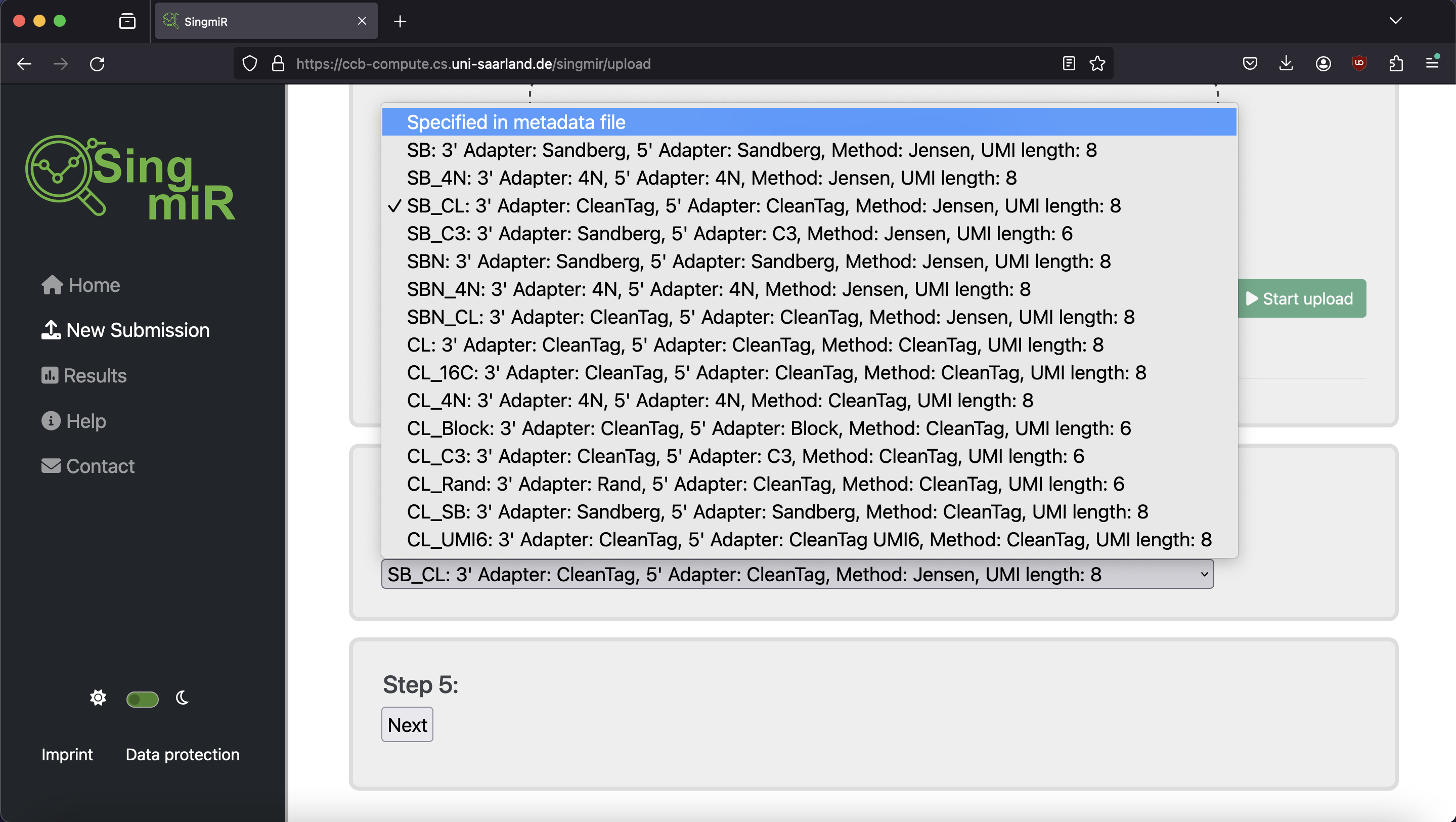
| Protocol variant | 3' adapter value | 5' adapter value | Method | UMI length |
|---|---|---|---|---|
| SB (Sandberg protocol I) | Sandberg | Sandberg | SB | 8 |
| SB_4N | 4N | 4N | SB | 8 |
| SB_CL | CleanTag | CleanTag | SB | 8 |
| SB_C3 | Sandberg | C3 | SB | 6 |
| SBN (Sandberg protocol II) | Sandberg | Sandberg | SBN | 8 |
| SBN_4N | 4N | 4N | SBN | 8 |
| SBN_CL | CleanTag | CleanTag | SBN | 8 |
| CL (CleanTag Protocol) | CleanTag | CleanTag | CleanTag | 8 |
| CL_16C | CleanTag | CleanTag | CleanTag | 8 |
| CL_4N | 4N | 4N | CleanTag | 8 |
| CL_Block | CleanTag | Block | CleanTag | 6 |
| CL_C3 | CleanTag | C3 | CleanTag | 6 |
| CL_Rand | Rand | CleanTag | CleanTag | 6 |
| CL_SB | Sandberg | Sandberg | CleanTag | 8 |
| CL_UMI6 | CleanTag | CleanTag UMI6 | CleanTag | 8 |
| 4N | 4n | 4N | 4N | 0 |
| 4N_C3 | 4N | C3 | 4N | 0 |
| 4N_CL | CleanTag | CleanTag | 4N | 0 |
| CATS | CATS | CATS | CATS | 0 |
Details
UMI length
Length of the UMI used for the library preparation method. For example, for the Sandberg protocol (Faridani et al., 2016, Hagemann-Jensen et al., 2018), the UMI was 8 nt long, while for the Sandberg 3’ CleanTag variant, the UMI was shortened to 6 nt.
5' Adapter
-
“Sandberg”: Adapters are following the methods of (Faridani et al., 2016, Hagemann-Jensen et al., 2018, denoted SB and SBN respectively. However, since the primary difference between the methods is the modification of technical variables other than the adapters, for the preprocessing, they are treated as one lable.
-
“CleanTag”: Modified adapters to inhibit formation of adapter dimers while allowing reverse transcriptase read-through (Shores et al., 2016).
-
“4N”: Adapter has 4 random nuclei at the ligation site, as per Giraldez et al., 2018).
-
“C3”: The 5’ adapter is complementary fo the 3’ adapter
-
“CleanTag UMI6”: The UMI of the 5′ adapter was shortened from 8 nt to 6 nt.
-
“Block”: 5’ CleanTag adapter replaced by an adapter with 3 additional uridine nucleotides at the 3’ end and an UMI shortened to 6 nt. The RT primer is altered, able to bind to adapter dimers.
-
“CATS”: Adapters corresponding to CATS small RNA sequencing kit for Illumina from Diogenode.
3' Adapter
-
“CleanTag”: Modified adapters to inhibit formation of adapter dimers while allowing reverse transcriptase read-through (Shores et al., 2016).
-
“Sandberg”: Adapters are following the methods of (Faridani et al., 2016, Hagemann-Jensen et al., 2018), denoted SB and SBN respectively. However, since the primary difference between the methods is the modification of technical variables other than the adapters, for the preprocessing, they are treated as one label.
-
“4N”: Adapter has 4 random nuclei at the ligation site, as per Giraldez et al., 2018.
-
“Rand”: CleanTag adapters with first 6 nucleotides exchanged with random nucleotides.
-
“CATS”: Adapters corresponding to CATS small RNA sequencing kit for Illumina from Diogenode
Method
-
“Jensen” : Corresponds to Hagemann-Jensen et al., 2018.
-
“Giraldez” : Corresponds to (Giraldez et al., 2018), primarily used in combination with 5’ adapter option C3.
References:
Faridani, O.R., et al (2016) Single-cell sequencing of the small-RNA transcriptome
Nature Biotechnology, 34(12), pp. 1264-1266, DOI: 10.1038/nbt.3701
Giraldez, M.D., et al (2018) Comprehensive multi-center assessment of small RNA-seq methods for quantitative MIRNA profiling
Nature Biotechnology, 36(8), pp. 746-757, DOI: 10.1038/nbt.4183
Hagemann-Jensen, M., et al (2018a) Small-seq for single-cell small-RNA sequencing
Nature Protocols, 13(10), pp. 2407-2424, DOI: 10.1038/s41596-018-0049-y
Hagemann-Jensen, M., et al (2018b) Small-seq for single-cell small-RNA sequencing
Nature Protocols, 13(10), pp. 2407-2424, DOI: 10.1038/s41596-018-0049-y
Hücker, S.M., et al (2021) Single-cell microrna sequencing method comparison and application to cell lines and circulating lung tumor cells
Nature Communications, 12(1), pp. 1264-1266, DOI: 10.1038/s41467-021-24611-w