Welcome to isomiRdb
A miRNA expression database with isoform resolution
isomiRdb stores miRNA and isomiR expression values for 42499 miRNA-seq samples collected from miRMaster, The Cancer Genome Atlas and Sequence Read Archive and uniformly processed from raw reads using sRNAbench . We cover over 70,000 different miRNA isoforms derived from miRBase (release 22.1) annotations.
Querying isomiRdb
isomiRdb can be queried in two three ways: sample query, miRNA query and isomiR query. Metadata from multiple sources has been gathered for all samples to allow specific querying using tissue, cell-line or cancer type.
Different visualisations are available for each type of query.
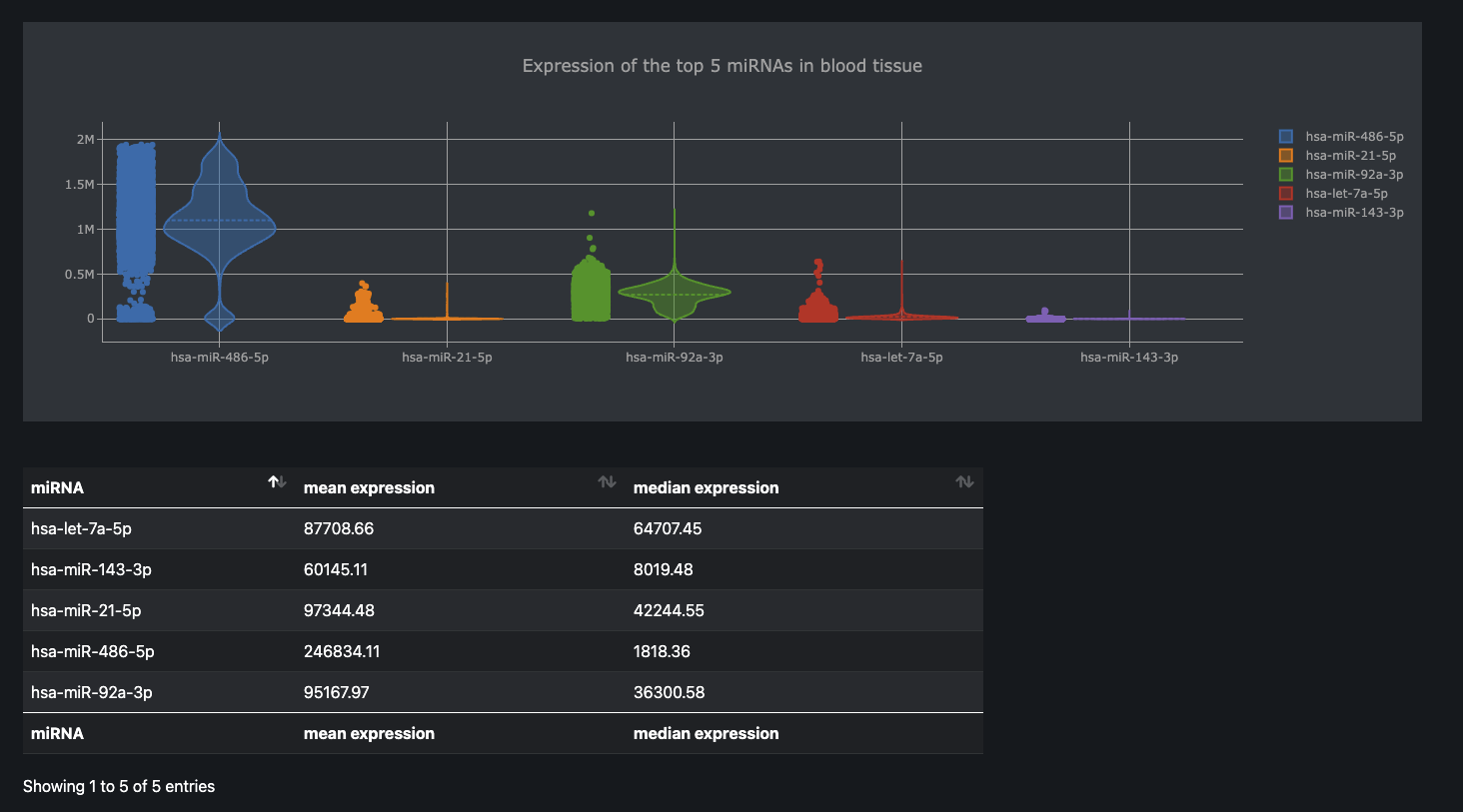
Sample query
Top miRNAs from each tissue hosted in the database can be queried and the resulting values can be downloaded.
Query samples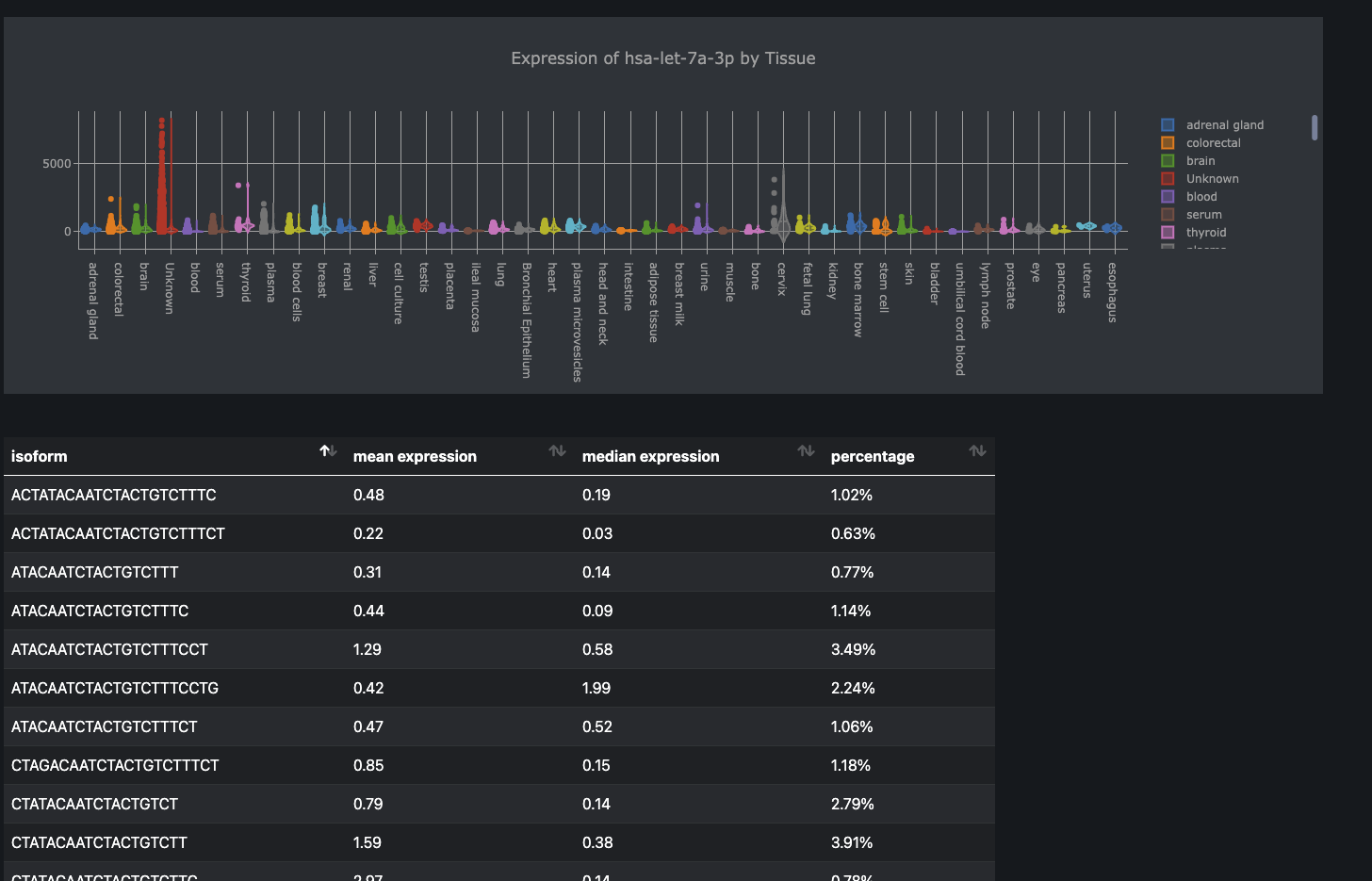
miRNA query
Individual miRNAs can be queried and visualized. The corresponding isomiR sequences and values are also displayed.
Query miRNAs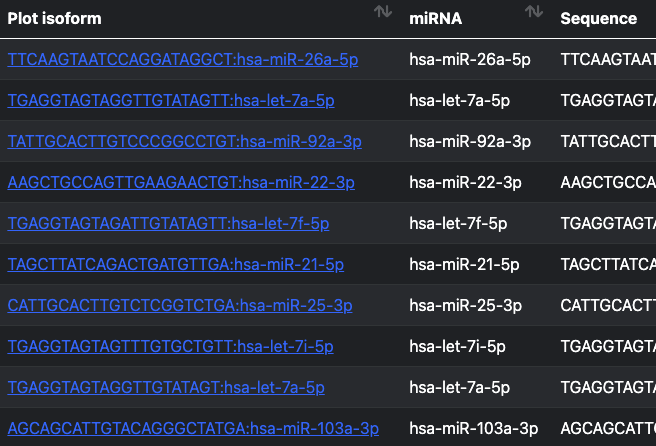
Query isomiRs
Over 3 million different isomiRs can be queried and visualized. If they didn't reach 1 RPM in at least 1% of the samples, no expression data is provided.
isomiR query